1. Introduction
This package provides methods to interact with the modelling framework Simplace1. Simplace is written
in Java (and some parts in Scala) so one can access it from R via rJava. The purpose of this package is to
simplify the interaction between R and Simplace, by providing functions to:
•initialize and configure Simplace
•load a simulation (solution and project)
•parameterize the simulation
•run whole simulation or run it stepwise
•get simulation output and convert it to formats suitable for R
2. Installing the Simplace Framework
For installing Simplace, please consult the webpage www.simplace.net.
A brief guide to install Simplace:
- If you don't have installed Java, please install an appropriate version of the (JRE or JDK) from java.com
- Download the console mode of Simplace from www.simplace.net
- Unpack the zip archive to your disk. You have to unpack the whole directory SIMPLACE and the directory must not be renamed
- Install the simplace package in R:
- Install.packages('simplace',repos=c('http://r-forge.r-project.org','http://cran.r-project.org'))
3. Basic Usage
The usage of Simplace in R follows roughly this scheme:
- init Simplace by providing the path to your simplace installation directory, your working directory and your outputs
- open a Simplace project form a solution (and project) le
- create a list of simulation parameters you want to change
- create and run a Simulation
- get the result from the simulation
- convert the result to a R object (data.frame, list etc.)
4. Troubleshooting
•Package rJava should be installed automatically with simplace. If not, install it manually: install.packages('rJava')
•Architecture of R and Java have to match. If you are using 64-bit Java, you have to use 64-bit R.
•If you want to use the development version instead of the console mode, make sure that the projects
simplace, lap, simplacerun and lapclient are in a common directory and set the installation dir to this directory.
5. Example
5.1 Run the simulation
> library(simplace)
> SimplaceInstallationDir <- "D:/java/simplace/"
> SimplaceWorkDir <- "D:/java/simplace/simplacerun/simulation/"
> SimplaceOutputDir <- "D:/java/simplace/simplacerun/output/"
> Solution <- "D:/java/simplace/simplacerun/simulation/gk/solution/complete/Complete.sol.xml"
> simplace <- initSimplace(SimplaceInstallationDir,SimplaceWorkDir,SimplaceOutputDir)
> openProject(simplace, Solution)
[1] "Java-Object{net.simplace.simulation.FWSimSession@2c9f9fb0}"
> parameter <- list()
> parameter$enddate <- "31-12-1992"
> sid <- createSimulation(simplace,parameter)
> runSimulations(simplace)
> result <- getResult(simplace,"DIAGRAM_OUT", sid);
> closeProject(simplace)
After specifying the directories and the solution, the framework is initialized and the project will be opened.
The end date of the simulation is (re)set and the simulation is run. After the run the result will be retrieved.
5.2 Get the result and plot it
> resf <- resultToDataframe(result)
> dates <- 300:730
> weights <- resf[dates, c("TOP_LINE_Roots","TOP_LINE_Leaves","TOP_LINE_Stems","TOP_LINE_StorageOrgans")]
> matplot(dates,weights,type="l",xlab="Days",ylab="Weight [g/m2]",main="Simulated Biomass")
> legend(300,800,legend=c("Roots","Leaves","Stems","Storage Organs"),lty=1:4,col=1:4)
Simulated Biomass
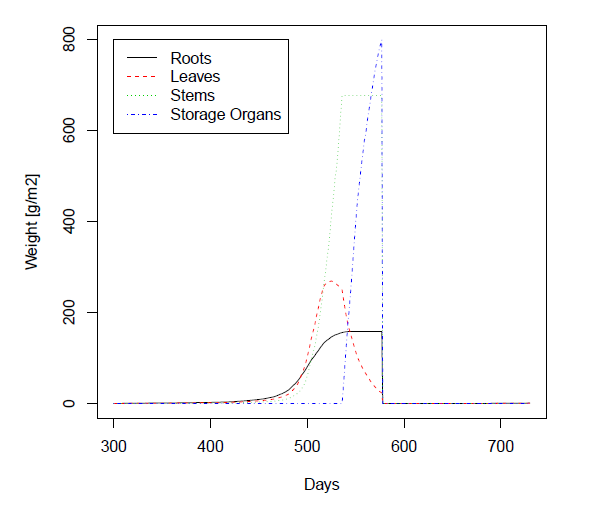
The result is converted to a dataframe. Interesting variables are extracted and then plotted.
5.3 Get arrays and plot them as contour plot
> resultlistexp <- resultToList(result,expand=TRUE)
> water <- resultlistexp$BOTTOM_ARRAY_VolumetricWaterContent
> wmat <- do.call(rbind,water)
> wmatpart <- wmat[dates,]
> layers <- dim(wmatpart)[2]
> filled.contour(dates,-(layers:1),wmatpart[,layers:1], xlab="Day", ylab="Layer", main="Water content in soil", color.palette = function(n){rgb((n:1)/n,(n:1)/n,1)})
Water content in soil
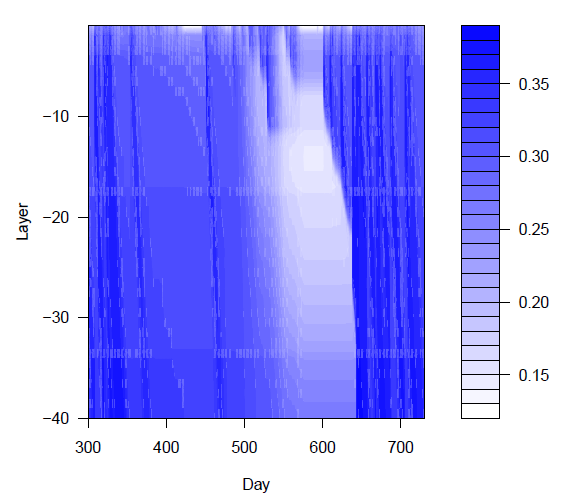
As the result contains an array which holds the water content for 40 layers, it is transformed to a list
and the array is expanded.
6. Wrapper client methods in R
Here you can find the list of the wrapper client methods with explanations.
simplacehandle <- initSimplace(InstallationDir,WorkDir,OutputDir) - starts Simplace, instead of InstallationDir one could use
a more generic parameter, e.g. Location, which could also be an URL it returns a handle to the running simplace instance which is used by subsequent functions.
openProject(simplacehandle, Solution, Project) - prepare a Session (formerly project) with associated solution and project file.
simid <- createSimulation(simplacehandle, parameterList, queue) - creates a Simulation and substitutes solution/project parameters
by the parameters in the parameterList; if queue is true, then the simulation is put into a queue, so one can run a couple of simulations at one time.
simlist <- getSimulationIDs(simplacehandle) - get the list of simulation ids of the current queue.
runSimulations(simplacehandle) - runs all the simulations in the queue.
result <- getResult(simplacehandle, outputId, simulationId) - fetches the result from the previous simulation run;
gets only the result for the specified simulation and the (memory) output id.
resultToDataframe(result, from, to), resultToList(result,expand,from, to) - converts the result to either a dataframe or a list.
Its possible to retrieve only the from to part of the result. If expand is true, then doublearrays are expanded to vectors, otherwise they remain just references to java objects.
If doublearrays are not needed, it saves time and space if not expanded.
resetSimulationQueue(simplacehandle) - resets the simulation queue.
closeProject(simplacehandle) - close the project.
runProject(simplacehandle) - runs the solution/project as defined in the original solution/projectfile; no data is kept in memory, it has to be fetched from the .csv outputs.
result<- stepSimulation(simplacehandle, count, filter) - runs the simulation stepwise; returns the whole varmap or only part of it defined by filter.
For example a calibration script in R would look like this:
simplace <- initSimplace(loc, wd, od)
openProject(simplace, mysolution)
ready <- false
parameters <- getSomeInitialParameters()
while (!ready)
{
id <- createSimulation(simplace,parameters,false)
runSimulations(simplace)
result <- getResult(simplace,"biomass",id)
df <- resultToDataframe(result)
parameters <- calculateSomehowParametersFromResults(df)
ready< <- isGoodEnough(df)
}
7. Further Material
There are a few talks and tutorials available from regular workshops and training about SIMPLACE in SVN repository simplace_doc:
•Interacting with SIMPLACE from R: Redmine link
•Advanced Calibration with R: Redmine link
•SIMPLACE in scripted workflows Redmine link